I. Brief Summary of Progress to Date:
The Yale efforts have been focused on two main areas: mapping transcribed regions throughout the ENCODE regions and mapping transcription factor binding sites. We plan to pursue each of these areas in the next year.
A. Mapping Transcribed Regions Throughout the ENCODE Regions
1. Platform Comparison
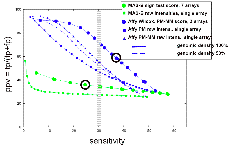
We spent considerable effort on platform comparison and mapping transcription using a variety of cell lines and tissues. We have compared the Affymetrix platform (25 b oligonucleotides) with the Maskless photolithography platform (36 b oligonucleotides) using two different standard hybridization conditions for the latter. These platforms involve different protocols and approaches. For example, Affymetrix involves probing with double stranded probes and has mismatched oligos on the array; maskless photolithography involves single stranded probes and does not use mismatches. A variety of scoring schemes were developed to compare these two platforms. Overall we found that based on detection of Gencode annotation, the Affymetrix platform has superior sensitivity and specificity compared to the maskless platform. For example, see Fig. 1 below. This is largely due to the increased numbers of features present on the Affymetrix arrays. As such we plan to utilize the Affymetrix platform for future transcription mapping studies. A manuscript describing these results is in press.
2. Transcription Maps
In collaboration with Affymetrix we also generated transcription maps for 7 different tissues and cell lines (all total RNA unless otherwise noted):
Cell lines:
NB4, NB4 + retinoic acid, NB4 + TPA, HeLaS3 (total and polyA+), BL2 Lymphoblast,
Tissues:
Placenta (polyA+), Neutrophils (from 11 different patients)
|
|
|
|
(LEFT) PPV versus sensitivity for two different ways of scoring the placenta Affy data, using 3 replicates (6 array features) unless otherwise stated: Wilcoxon signed rank test (blue circles), and standard sign test (using PM-MM values: cyan triangles ; using PM-MM-only values: values; yellow squares using PM values only). The result from reducing the genomic density of the Affy array to 50% (ie., removing the data from every second probe) is also shown, using PM-MM values (3 replicates: cyan triangles, dashed line; single replicate only: grey triangles, dashed line). (RIGHT) PPV versus sensitivity for MAS-B and Affy placenta data, varying the segmentation threshold from 70th percentile (to the right in the figure) to 99th percentile (to the left). The average results of TARs generated from raw intensities from single arrays for Affy (PM only (blue squares), and PM-MM (blue triangles; solid line actual genomic density;, dashed line 50% genomic density)) and MAS-B (green squares) are plotted, as well as scored results for Affy (blue circles) and MAS-B (green circles). |
|
These results represent a sizable fraction of the transcription map data present in ENCODE. We found a) many novel transcribed regions; the number depends upon the threshold used. b) neutrophils exhibit more extensive transcription in some apparently unannotated regions relative to other RNAs and c) surprisingly, variation among cell lines is greater than that among individuals.